Antimicrobial Peptides
- Details
- Last Updated: Friday, 14 July 2023 09:28
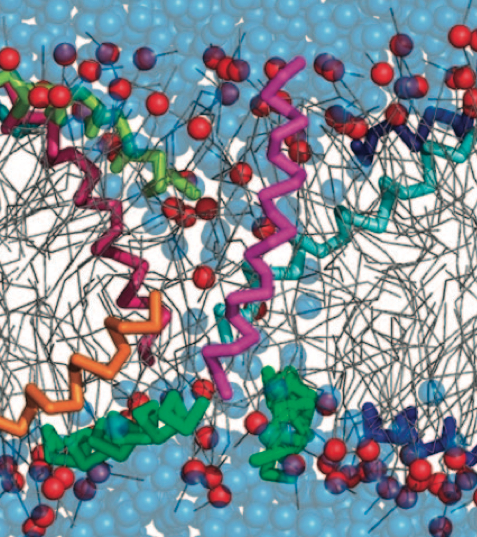 Antimicrobial peptides (AMPs) are currently in the spotlight as potential candidates to overcome bacterial resistance to conventional antibiotics. These peptides are natural weapons produced by a variety of organisms, including insects, animals and plants. How AMPs kill bacteria by interacting with the cell membrane is not fully understood. These peptides, often small and cationic, are secreted into the aqueous phase and bind quickly to the target membrane. At a certain threshold concentration, AMPs permeabilize the membrane, either by forming a discrete pore or by disrupting the bilayer structure.
Antimicrobial peptides (AMPs) are currently in the spotlight as potential candidates to overcome bacterial resistance to conventional antibiotics. These peptides are natural weapons produced by a variety of organisms, including insects, animals and plants. How AMPs kill bacteria by interacting with the cell membrane is not fully understood. These peptides, often small and cationic, are secreted into the aqueous phase and bind quickly to the target membrane. At a certain threshold concentration, AMPs permeabilize the membrane, either by forming a discrete pore or by disrupting the bilayer structure.
With the Martini model it is in principle possible to simulate the AMP induced membrane poration. A first example was shown in [1, see figure], with a toroidal pore stabilized by magainin peptides resembling pores seen with atomistic models. Simulation of spontaneous pore formation has also proven possible, however, only in the case of very thin bilayers [2], at very high concentrations [8], or with asymmetric bilayers [10]. Other attempts to simulate AMP induced membrane poration also reveal potential problems with Martini in this respect, such as formation of dehydrated pores [3,4] or membrane buckling [5].
It appears that the formation of pores in lipid bilayers costs more energy with Martini than with more detailed force fields [6], which might be related to the implicit screening of charges in the Martini force field. With the polarizable water model, better results are obtained [6], but in general one needs to be cautious using Martini for simulations in which polar or charged particles need to penetrate a hydrophobic enrironment such as the membrane interior.
The ability of AMPs in a surface bound state to cluster charged lipids does not suffer from this problem, and is therefore expected to be a real phenomenon well suited for Martini [7,11,13]. Likewise, Martini allows for detailed calculations of the binding free energy of AMPs [9], as well as their preferred sorting betwnee different membrane environments [12].
- [1] L. Monticelli, S.K. Kandasamy, X. Periole, R.G. Larson, D.P. Tieleman, S.J. Marrink.The MARTINI coarse grained forcefield: extension to proteins. JCTC, 4:819-834, 2008. abstract
- [2] A. Rzepiela, D. Sengupta, N. Goga, S.J. Marrink. Membrane poration by antimicrobial peptides combining atomistic and coarse grained descriptions. Farad. Discuss., 144:431-443, 2010. abstract
- [3] L. Thogersen, B. Schiott, T. Vosegaard, N.C. Nielsen, E. Tajkhorshid. Peptide aggregation and pore formation in a lipid bilayer: a combined coarse-grained and all atom molecular dynamics study. Biophys. J. 95:4337-4347, 2008.
- [4] P.J. Bond, D.L. Parton, J.F. Clark, M.S.P. Sansom. Coarse-grained simulations of the membrane-active antimicrobial Peptide maculatin 1.1. Biophys J. 95:3802-15, 2008.
- [5] H.J. Woo, A. Wallqvist. Spontaneous buckling of lipid bilayer and vesicle budding induced by antimicrobial peptide magainin 2: a coarse-grained simulation study. J. Phys. Chem. 115:8122-8129, 2011.
- [6] W.F.D. Bennett, D.P. Tieleman. Defect and pore formation in atomistic and coarse-grained lipid membranes: pushing the limits of coarse graining, J. Chem. Theo. Comp., 7:2981-2988, 2011.
- [7] A.A. Polyansky, R. Ramaswarny, P.E. Volynsky, I.F. Sbalzarini, S.J. Marrink, R.G. Efremov. Antimicrobial peptides induce growth of phosphatidylglycerol domains in a model bacterial membrane. J. Phys. Chem. Letters, 1:3108–3111, 2010. abstract
- [8] M.L. Berkowitz, K. Santo. The difference between Magainin-2 and Melittin assemblies in phosphatidylcholine bilayers: results from coarse-grained simulations. J. Phys. Chem. B, Just Accepted Manuscript, 2012. DOI:10.1021/jp212018f
- [9] C.I.E. von Deuster, V. Knecht. Competing interactions for antimicrobial selectivity based on charge complementarity. BBA Biomembr., 1808:2867-287, 2011.
- [10] Z. Li, H. Ding, Y. Ma. Translocation of polyarginines and conjugated nanoparticles across asymmetric membranes. Soft Matter, ASAP, 2012. DOI: 10.1039/c2sm26519b
- [11] J.N. Horn, J.D. Sengillo, D. Lin, T.D. Romo, A. Grossfield. Characterization of a potent antimicrobial lipopeptide via coarse-grained molecular dynamics BBA Biomembr. 1818:212-218, 2012.
- [12] J. Su, S.J. Marrink, M.N. Melo. Localization Preference of Antimicrobial Peptides on Liquid-Disordered Membrane Domains. Front. Cell Dev. Biol., 8:350, 2020. doi.org/10.3389/fcell.2020.00350
- [13] A. Melcrova, S. Maity, J. Melcr, N.A.W. de Kok, M. Gabler, J. van der Eyden, W. Stensen, J.S M. Svendsen, A.J.M. Driessen, S.J. Marrink, W.H. Roos. Lateral membrane organization as target of an antimicrobial peptidomimetic compound. Nature Commun. 4, 4038, 2023. doi:10.1038/s41467-023-39726-5


























































































































































