Membrane shapes
- Details
- Last Updated: Wednesday, 25 January 2023 14:26
Membranes provide identity not only to the cell as a whole, through the enveloping plasma membrane, but also to many internal organelles. Membrane spatial organization ranges from a locally flat, lamellar geometry to highly curved ones as can be found, for instance, in the endoplasmic reticulum, Golgi apparatus, thylakoids, or mitochondria. Moreover, these structures are dynamic, involved in many remodelling processes often with highly curved intermediates.
Martini-based simulations have proven useful in capturing these membrane shapes, from the strongly curved regions present in small lipid vesicles [1,2] and lipid tethers [3], to the large scale undulations of planar membrane patches. These type of simulations provide important clues about the interplay between curvature and protein/lipid sorting (see Figure, taken from [4]).
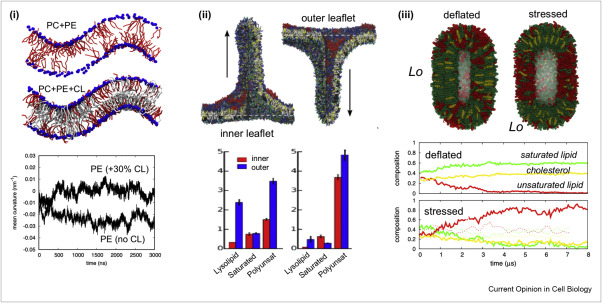
Recently, efforts are directed in modeling realistic shapes of large cellular substructures and even entire organelles, such as the mitochondrion [5]. A sophisticated tool to generate Martini membranes with arbitrary curvatures is TS2CG [5,6], and a tool to identify lipid leaflets in complex curved membranes is also available [7].
In addition, recent Martini-based simulation efforts are directed to understand membrane curvature generation via protein-lipid interplay. Recent examples include the strong membrane deformations induced by ECF transporters [8] and around a signal peptidase [9], and membrane deformation induced by functionalized oligonucleotides [10].
- [1] H.J. Risselada, S.J. Marrink. Curvature effects on lipid packing and dynamics in liposomes revealed by coarse grained molecular dynamics simulations. Phys. Chem. Chem. Phys., 11:2056-2067, 2009.
- [2] H.J. Risselada, S.J. Marrink, M. Muller. Curvature-dependent elastic properties of liquid-ordered domains result in inverted domain sorting on uniaxially compressed vesicles. Phys. Rev. Lett., 106:148102, 2011. abstract
- [3] S. Baoukina, H.I. Ingólfsson, S.J. Marrink, D.P. Tieleman. Curvature‐Induced Sorting of Lipids in Plasma Membrane Tethers. Advanced Theory Simul., 1:1800034, 2018. doi:10.1002/adts.201800034
- [4] W. Pezeshkian, S.J. Marrink. Simulating realistic membrane shapes. Curr. Opin. Cell Biol., 71:103-111, 2021. https://doi.org/10.1016/j.ceb.2021.02.009
- [5] W. Pezeshkian, M. Konig, T.A. Wassenaar, S.J. Marrink. Backmapping triangulated surfaces to coarse-grained membrane models. Nature Commun. 11:2296, 2020. doi.org/10.1038/s41467-020-16094-y
- [6] W. Pezeshkian, M. König, S.J. Marrink, J.H. Ipsen. A multi-scale approach to membrane remodeling processes. Front. Mol. Biosci. 6, 59, 2019. doi:10.3389/fmolb.2019.00059
- [7] B.M.H. Bruininks, A.S. Thie, P.C.T. Souza, T.A. Wassenaar, S. Faraji, S.J. Marrink. Sequential Voxel-Based Leaflet Segmentation of Complex Lipid Morphologies. J. Chem. Theory Comput., 2021. online
- [8] I. Faustino, H. Abdizadeh, P.C.T. Souza, A. Jeucken, W.K. Stanek, A. Guskov, D.J. Slotboom, S.J. Marrink. Membrane mediated toppling mechanism of the folate energy coupling factor transporter. Nature Commun. 11:1763, 2020. doi.org/10.1038/s41467-020-15554-9
- [9] A.M. Liaci, ..., S.J.Marrink, R.A.Scheltema, F. Förster. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell. 81, 3934-3948, 2021. https://doi.org/10.1016/j.molcel.2021.07.031
- [10] N. De Franceschi, W. Pezeshkian, A. Fragasso, B.M.H. Bruininks, S. Tsai, S.J. Marrink, C. Dekker. Synthetic Membrane Shaper for Controlled Liposome Deformation. ACS Nano, online, 2023. doi:10.1021/acsnano.2c06125


























































































































































