Protein-Lipid Fingerprints
- Details
- Last Updated: Friday, 14 July 2023 09:31
 It becomes more and more clear that protein-lipid interactions are of fundamental importance to the (mal)functioning of membrane embedded or associated proteins. Both specific binding (lipid binding sites) as well as the non-specific binding (general preference to reside in certain membrane environments) are molecular mechanisms that can steer the behavior of proteins either directly or indirectly.
It becomes more and more clear that protein-lipid interactions are of fundamental importance to the (mal)functioning of membrane embedded or associated proteins. Both specific binding (lipid binding sites) as well as the non-specific binding (general preference to reside in certain membrane environments) are molecular mechanisms that can steer the behavior of proteins either directly or indirectly.
The Martini model has been succesfully applied to simulate protein-lipid interactions, with early examples showing how lipid membrane composition affects the self-assembly of GPCRs [1], glycophorin [2] ,and more [3,4], or the gating behavior of mechanosenstive channels [5]. More recent examples include the salt-dependent binding of actin filaments to charged membranes [17] and the lipid-dependent toppling mechanism of the energy coupling tranpsort factor [16].
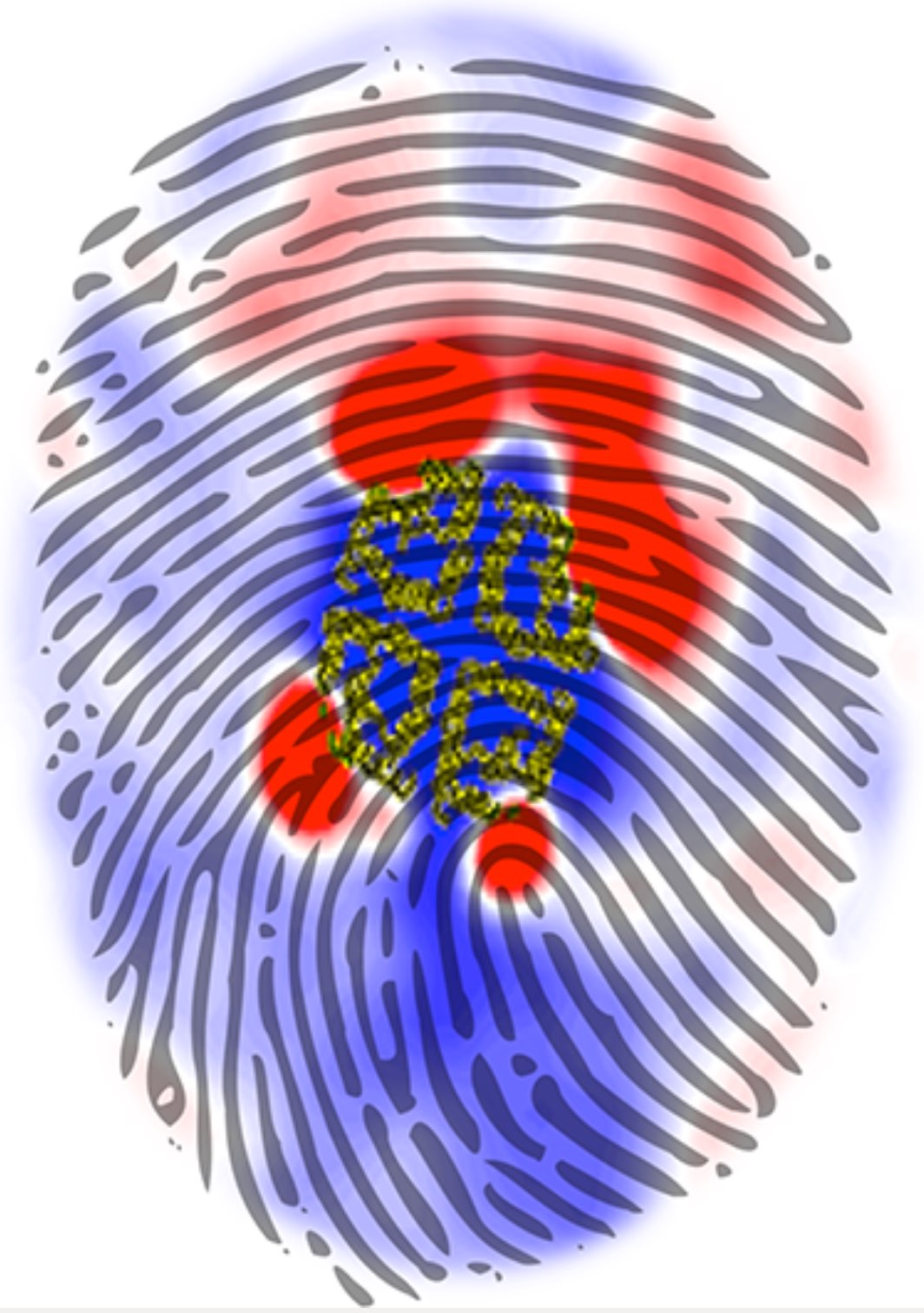
Over the past decade, many more specific lipid binding sites have been resolved based on Martini simulations, including binding sites for cardiolipins [6,7], gangliosides [8], galactolipids [9,10], PIPs [11,12,20,21], ceramides [13], anionic lipids [14], cholesterol [15], and many more - recently reviewed in [19]. A recent development is to study protein-lipid interactions in more complex environments, such as the plasma membrane, giving rise to the concept of protein-lipid fingerprints [18, see Figure].
[1] X. Periole, T. Huber, S.J. Marrink, T. P. Sakmar. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. JACS, 129:10126-10132, 2007. abstract
[2] D. Sengupta, S.J. Marrink. Lipid mediated Interactions tune the association of Glycophorin A helix and its disruptive mutants in membranes. Phys. Chem. Chem. Phys., 12:12987-12996, 2010. abstract
[4] D.H. de Jong, C.A. Lopez, S.J. Marrink. Molecular view on protein sorting into liquid-ordered membrane domains mediated by gangliosides and lipid anchors. Farad. Discuss., 161:347-363, 2013. abstract
[5] N. Mukherjee, M.D. Jose, J.P. Birkner, M. Walko, H.I. Ingólfsson, A. Dimitrova, C. Arnarez, S.J. Marrink, A. Koçer. The activation mode of the mechanosensitive ion channel, MscL, by lysophosphatidylcholine differs from tension-induced gating. FASEB J., 28:4292-4302, 2014
[6] C. Arnarez, J.P. Mazat, J. Elezgaray, S.J. Marrink, X. Periole. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. JACS, 135:3112–3120, 2013. open access
[7] C. Arnarez, S.J. Marrink, X. Periole. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci. Rep., 3:1263, 2013. open access
[8] R.X. Gu, H.I. Ingólfsson, A.H. de Vries, S.J. Marrink, D.P. Tieleman. Ganglioside-lipid and ganglioside-protein interactions revealed by coarse-grained and atomistic molecular dynamics simulations. JPCB, 121:3262–3275, 2017. open access
[9] F.J. van Eerden, M.N. Melo, P.W.J.M. Frederix, S.J. Marrink. Prediction of thylakoid lipid binding sites on photosystem II. Biophys. J. 113:2669-2681, 2017. open access
[10] S. Thallmair, P.A. Vainikka, S.J. Marrink. Lipid Fingerprints and Cofactor Dynamics of Light-Harvesting Complex II in Different Membranes. Biophys. J., 116:1446-1455, 2019. doi:10.1016/j.bpj.2019.03.009
[11] F. Sun, C.F.E. Schroer, L. Xu, H. Yin, S.J. Marrink, S.Z. Luo. Molecular Dynamics of the Association of L-Selectin and FERM Regulated by PIP2. Biophys. J., 114:1858–1868, 2018. doi:10.1016/j.bpj.2018.02.034
[12] F. Sun, C.F.E. Schroer, C.R. Palacios, L. Xu, S.Z. Luo, S.J. Marrink. Molecular mechanism for bidirectional regulation of CD44 for lipid raft affiliation by palmitoylations and PIP2. PLoS Comput. Biol. 16:e1007777, 2020. doi.org/10.1371/journal.pcbi.1007777
[13] S. Dadsena, S. Bockelmann, J.G.M. Mina, D.G. Hassan, S. Korneev, G. Razzera, H. Jahn, P. Niekamp, D. Müller, M. Schneider, F.G. Tafesse, S.J. Marrink, M.N. Melo, J.C.M. Holthuis, Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nature Commun. 10:1832, 2019. doi:10.1038/s41467-019-09654
[14] S. Koch, `M. Exterkate, C.A. López, M. Patro, S.J. Marrink, A.J.M. Driessen Two distinct anionic phospholipid-dependent events involved in SecA-mediated protein translocation. BBA-Biomembr. 1861, 183035, 2019. doi.10.1016/j.bbamem.2019.183035
[15] A. Buyan, C.D. Cox, J. Barnoud, J. Li, H.S.M. Chan, B. Martinac, S.J. Marrink, B. Corry. Piezo1 forms specific, functionally important interactions with phosphoinositides and cholesterol. Biophys. J. 119:1683-1697, 2020. doi.10.1016/j.bpj.2020.07.043
[16] I. Faustino, H. Abdizadeh, P.C.T. Souza, A. Jeucken, W.K. Stanek, A. Guskov, D.J. Slotboom, S.J. Marrink. Membrane mediated toppling mechanism of the folate energy coupling factor transporter. Nature Commun. 11:1763, 2020. doi.org/10.1038/s41467-020-15554-9
[17] C.F.E. Schroer, L. Baldauf, L. van Buren, T.A. Wassenaar, M.N. Melo, G. Koenderink, S.J. Marrink. Charge-dependent interactions of monomeric and filamentous actin with lipid bilayers. PNAS,
[18] V. Corradi, E. Mendez-Villuendas, H.I. Ingólfsson, R.X. Gu, I. Siuda, M.N. Melo, A. Moussatova, L.J. DeGagné, B.I. Sejdiu, G. Singh, T.A. Wassenaar, K. Delgado Magnero, S.J. Marrink, D.P. Tieleman. Lipid–Protein Interactions Are Unique Fingerprints for Membrane Proteins. ACS Central Science 4:709–717, 2018. doi:10.1021/acscentsci.8b00143
[19] V. Corradi, B.I. Sejdiu, H. Mesa-Galloso, H. Abdizadeh, S.Y. Noskov, S.J. Marrink, D.P. Tieleman. Emerging Diversity in Lipid–Protein Interactions, Chem. Review, 119:5775–5848, 2019. doi:10.1021/acs.chemrev.8b00451
[20] V. Thallmair, L. Schultz, W. Zhao, S.J. Marrink, D. Oliver, S. Thallmair. Two cooperative binding sites sensitize PI(4,5)P2 recognition by the tubby domain. Science Advances 8 (36), eabp9471, 2022. DOI: 10.1126/sciadv.abp9471
[21] M. Ren, L. Zhao, Z. Ma, H. An, S.J. Marrink, F. Sun. Molecular basis of PIP2-dependent conformational switching of phosphorylated CD44 in binding FERM. Biophys. J., 122 (13), 2675-2685, 2023. doi:10.1016/j.bpj.2023.05.021


























































































































































