Nucleic Acids
Deoxyribonucleic acid (DNA)
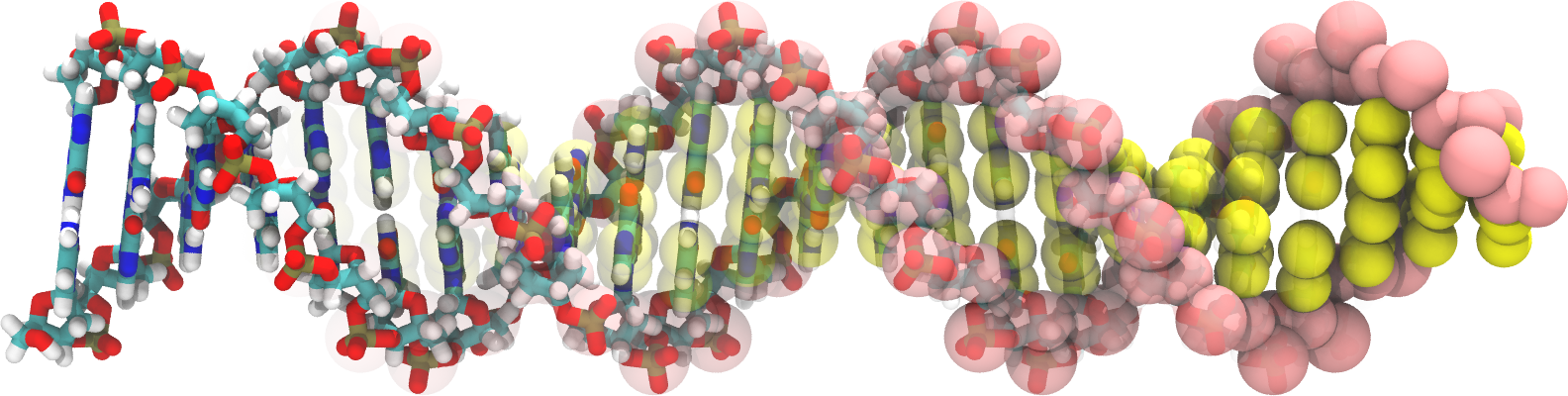
Martini 2 DNA parameters are available below including a martinize-dna script to generate Martini topology files for DNA molecules. In addition, an example of an all atomistic 24 bp dsDNA in .pdb format, together with its course grained mapping and bonded parameters, using the stiff elastic network settings is provided here. For further background reading on the Martini 2 DNA parameters, see:
- J.J. Uusitalo, H.I. Ingólfsson, P. Akhshi, D.P. Tieleman, S.J. Marrink. Martini coarse-grained force field: extension to DNA. JCTC, (doi: 10.1021/acs.jctc.5b00286)
References to these works can be found in the headers of the corresponding .itp files.
| Version | Description |
|---|---|
| martini-dna-150909.tar | martini_v2.1P-dna.itp: Fixed missing T-bead - S-bead interactions. |
| martini-dna-150817.tar | dna_backmapping_files: Fixed a bug in CHARMM36 thymine files. |
| martini-dna-150814.tar | martinize-dna.py: Changed the default cut-off for separating chains to 4 Angstroms. |
| martini-dna-150722.tar | Original release |
Ribonucleic acid (RNA)
The Martini 2 RNA parameters are available as a package with a martinize script to generate the Martini 2 topology files for RNA molecules. It also includes a backmapping file to convert the coarse RNA resolution into atomistic RNA (AMBER and CHARMM) using the backmapping procedure described here. In addition, a tutorial to start running your first Martini 2 RNA system is available here. For further background reading on the Martini 2 DNA parameters, see:
- J.J. Uusitalo, H.I. Ingólfsson, S.J. Marrink, I. Faustino. Martini Coarse-Grained Force Field: Extension to RNA. Biophys. J., (doi: 10.1016/j.bpj.2017.05.043)
References to these works can be found in the headers of the corresponding .itp files.
| Version | Description |
|---|---|
| na-tutorials_20170815.tar | Original release |