Announcing the 2015 Martini Coarse-Graining Workshop
- Details
- Last Updated: Wednesday, 12 August 2015 09:47
Are you impressed by the interesting science done by using the Martini coarse-grained model, and do you want to hear from the developers about the philosophy, back-ground, current, and future developments of Martini? Moreover, do you want to practice coarse-graining skills under the guidance of Martini experts? The Martini summerschool is the place for you!
REGISTRATION IS NOW CLOSED - WE ARE FULL!
Read more: Announcing the 2015 Martini Coarse-Graining Workshop




























































































































































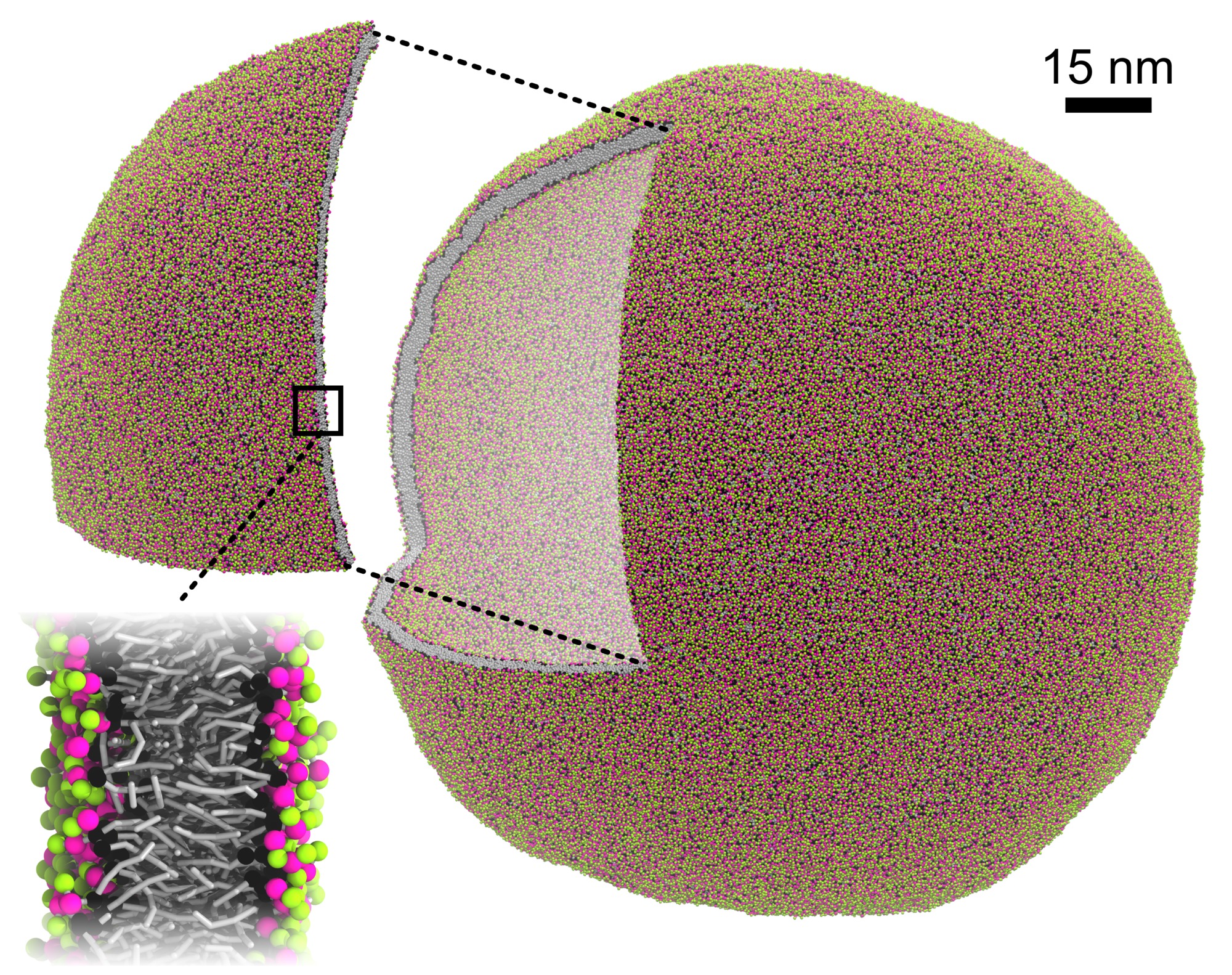
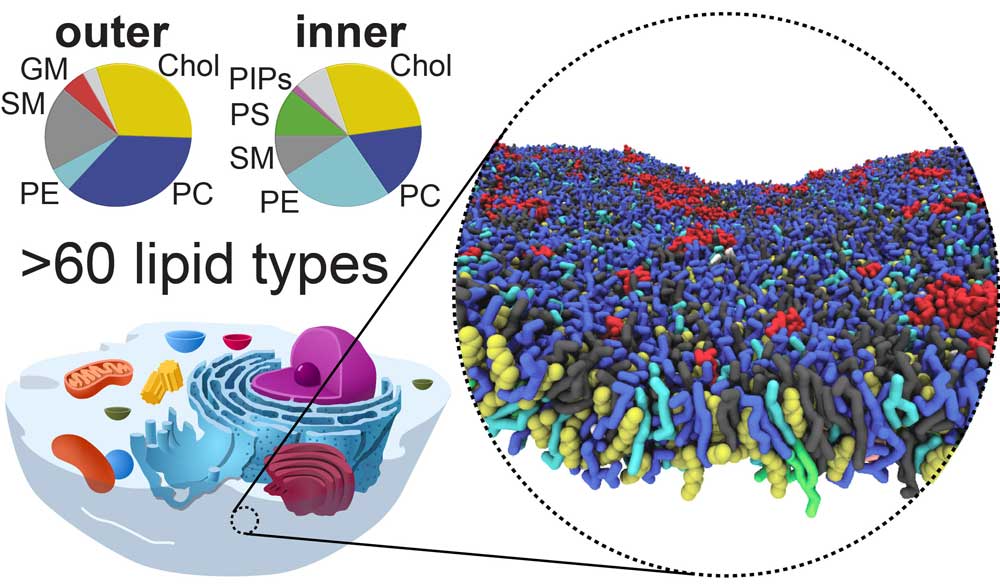 Plasma membrane model composed of 60+ lipid types
Plasma membrane model composed of 60+ lipid types