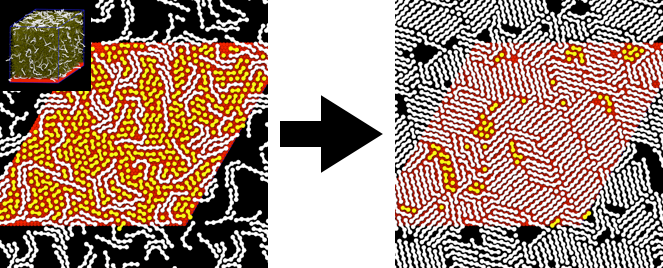
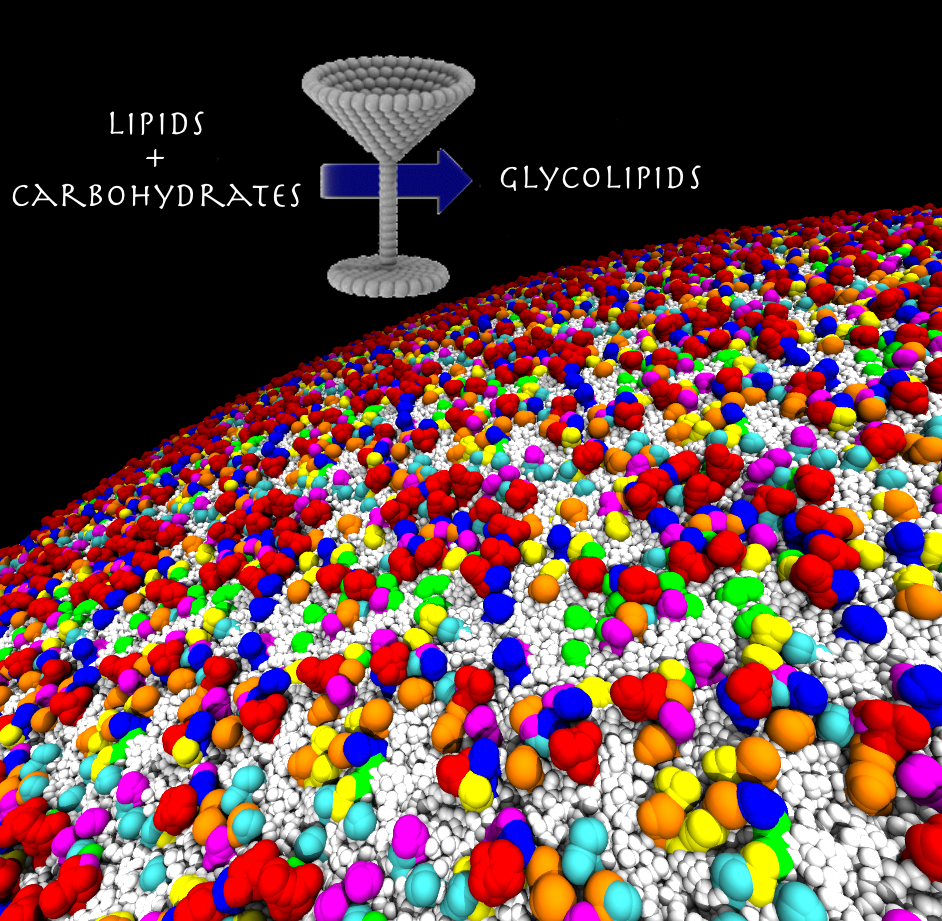
Website updated
- Details
- Last Updated: Saturday, 09 November 2013 13:08
There was a major update on the cgmartini.nl website on August 22nd. Due to the update and a move to a new server, a handful of forum posts between August 20-22 were lost. We are very sorry for the inconvenience.