Whole-cell Martini model
- Details
- Last Updated: Wednesday, 18 January 2023 08:34
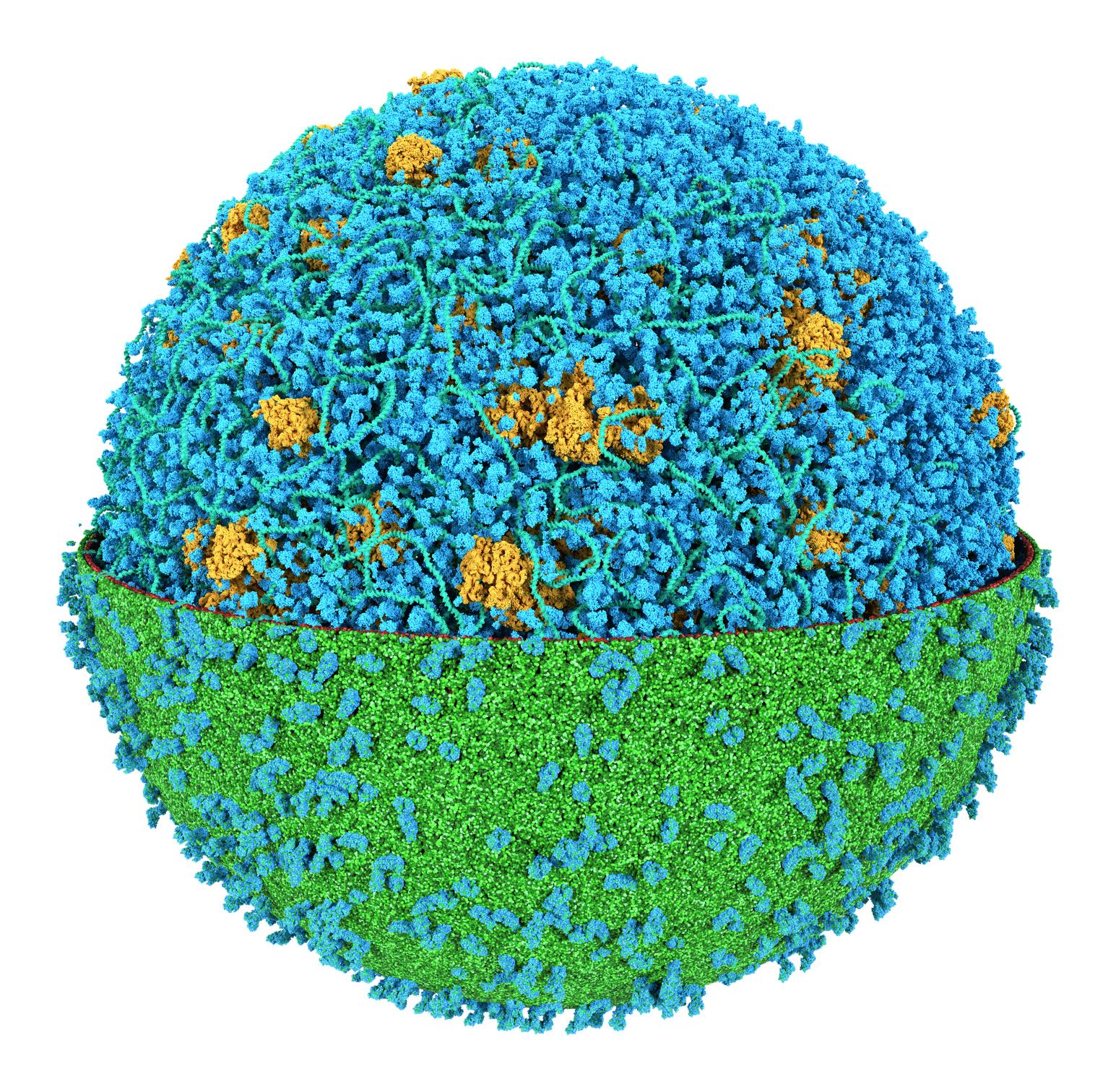
We got him ! Together with the group of Zan Luthey-Schulten we modeled an entire cell (JCVI-Syn3a) at the Martini level of resolution. Featuring more than 60,000 proteins, millions of lipids and metabolites, 500 ribosomes and a 500 kbp circular dsDNA.
For details, read our perspective paper: Stevens et al., 'Molecular dynamics simulation of an entire cell', Frontiers in Chemistry, vol 11, 2023. https://doi.org/10.3389/fchem.2023.1106495



























































































































































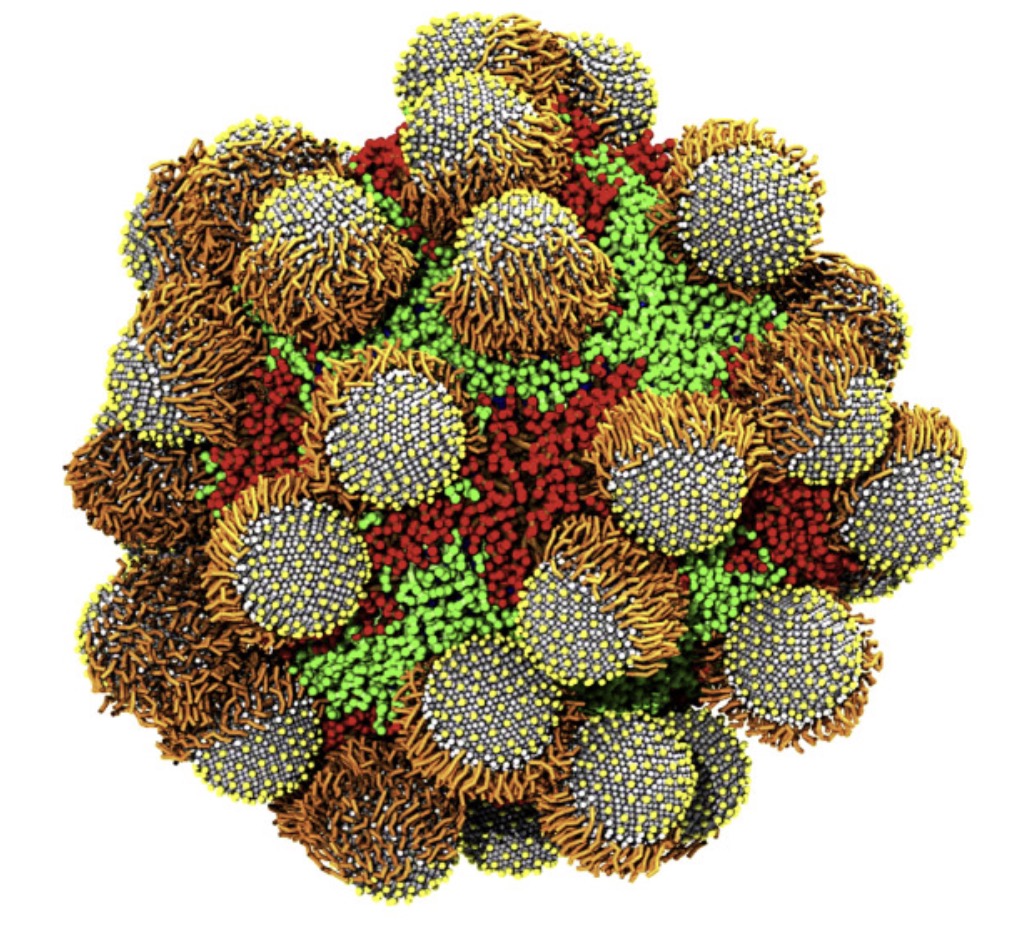

 Recently, two parameter sets for modeling carbohydrates with Martini 3 were published in JCTC.
Recently, two parameter sets for modeling carbohydrates with Martini 3 were published in JCTC.