Deep eutectic solvents
- Details
- Last Updated: Thursday, 16 December 2021 13:10
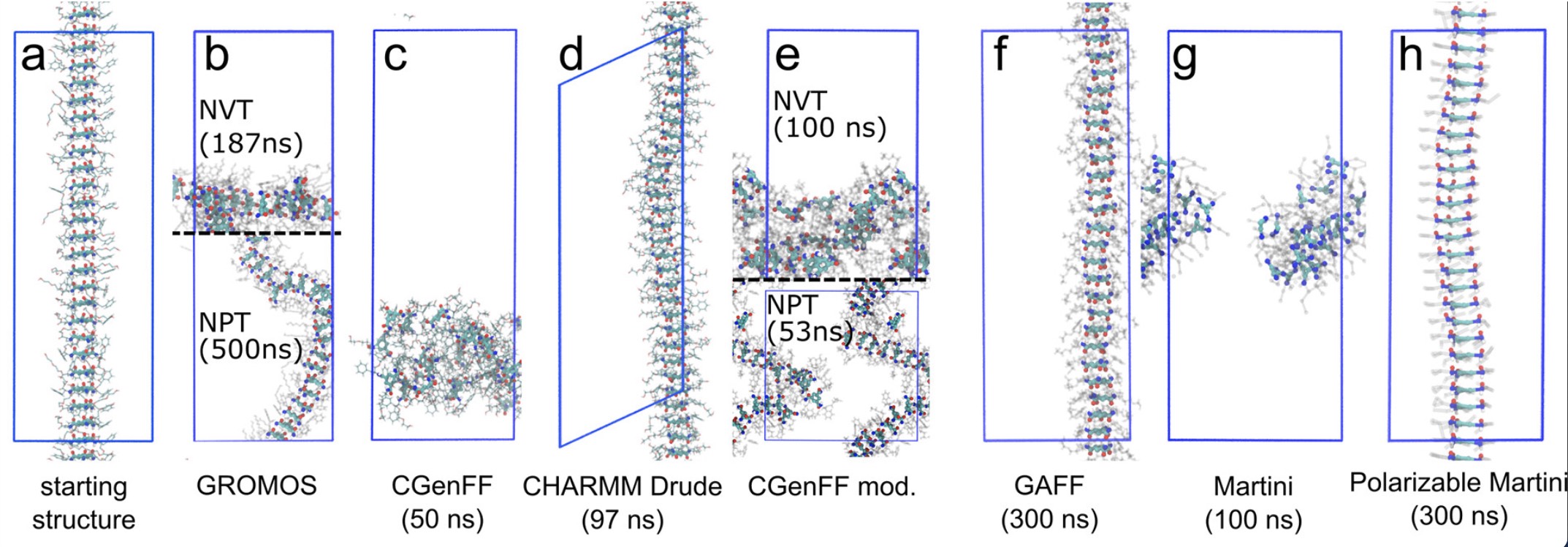
 Deep eutectic solvents (DESs) are a more environmentally friendly, cost-effective, and recyclable alternative for ionic liquids. Since the number of possible deep eutectic solvents is very large, there are needs for effective methods to predict the physicochemical nature of possible new deep eutectic solvents that are not met by the currently available models.
Deep eutectic solvents (DESs) are a more environmentally friendly, cost-effective, and recyclable alternative for ionic liquids. Since the number of possible deep eutectic solvents is very large, there are needs for effective methods to predict the physicochemical nature of possible new deep eutectic solvents that are not met by the currently available models.
To meet this challenge, we parameterized and validated a first set of DESs compatible with Martini 3, and showed its application in simulating liquid-liquid extraction processes.
For details, check Vainikka et al., ACS Sust. Chem. Engin., online.