Moltemplate
- Details
- Last Updated: Thursday, 11 February 2021 15:43
Want to do fancy simulations ? Perhaps Moltemplate is a suitable tool - it can now generate input files which allow you to run Martini simulations for complex systems using LAMMPS.
 General Purpose Coarse-Grained Force Field
General Purpose Coarse-Grained Force Field 

























































































































































Want to do fancy simulations ? Perhaps Moltemplate is a suitable tool - it can now generate input files which allow you to run Martini simulations for complex systems using LAMMPS.
A cool example of validating Martini in capturing the phase behaviour of wax-water mixtures under confinement, fundamental to industiral processes such as the synthesis of transportation fuels. For details, see this paper from Economou and coworkers.
We have job openings for two positions:
(1) PhD position in Non-Equilibrium Cellular Processes
(2) PhD/Postdoc position in Biomolecular Simulation of Photosynthetic Membranes
For more details on these positions, and how to apply, see here.
 Be careful - drinking too many Martinis or being an inexperienced drinker may cause a severe hangover.
Be careful - drinking too many Martinis or being an inexperienced drinker may cause a severe hangover.
A recent paper from the Voth group provides a clear example of this downside, in particular the entropy-enthalpy compensation which is inherent to CG models in general, including Martini.
For details, see: Jarin et al., JCTC, 2021.
Check out this nice paper on modeling E.coli lipid membranes at multiple levels of compositional complexity, showing that the presence of many different lipid types gives these membranes their distinct properties. And, demonstrating excellent performance of the preliminary Martini 3 lipids !
Pluhackova & Horner, "Native-like membrane models of E. coli polar lipid extract shed light on the importance of lipid composition complexity", BMC Biology, 2021. online.
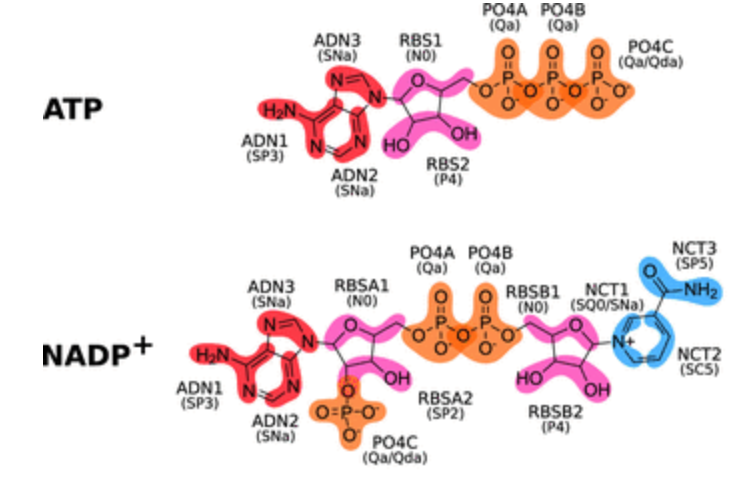
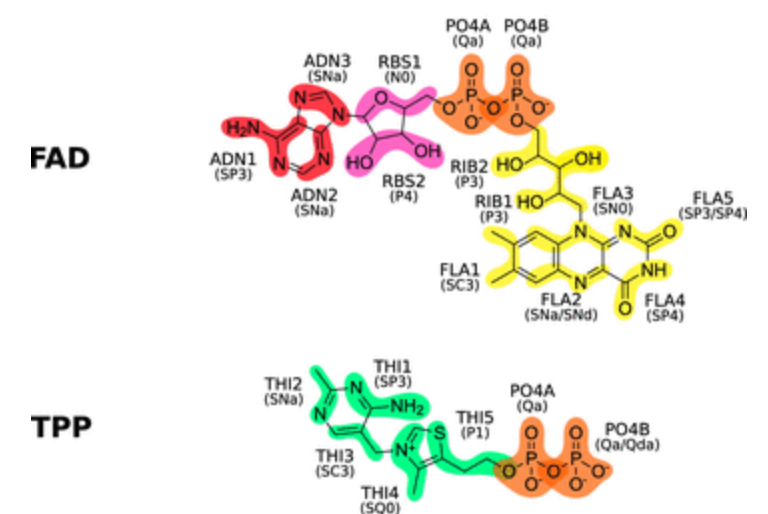
All these small but essential molecules Martinized especially for you by the Melo team. Check it out: Sousa et al., JCIM, 2021
The paper describes Martini topologies for key metabolites and cofactors (FAD, FMN, riboflavin, NAD, NADP, ATP, ADP, AMP, and thiamine pyrophosphate) in different oxidation and protonation states as well as smaller molecules derived from them (among others, nicotinamide, adenosine, adenine, ribose, thiamine, and lumiflavin), summing up a total of 79 different molecules.
The topologies are available for download here!