Death is just the beginning ....
- Details
- Last Updated: Saturday, 24 October 2020 08:52
Check out a nice paper from Lucendo et al, in PNAS, showing how TM helix dimerization plays a role in apoptosis. Featuring the Martini 3 beta release, corroborated by atomistic simulations and experimental data.



























































































































































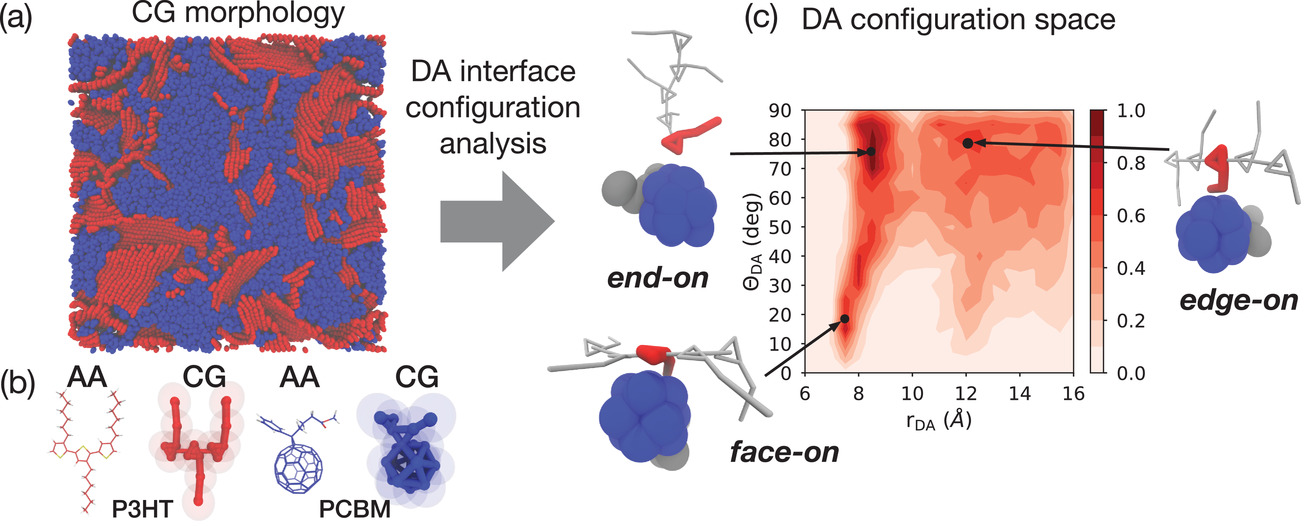 Martini is not only suitable to simulate biomolecular processes, but also increasingly to study material properties. Check the latest paper from our group on resolving the impact of polar side chains on electronic and structural properties of donor-acceptor (DA) interfaces of organic semiconductors:
Martini is not only suitable to simulate biomolecular processes, but also increasingly to study material properties. Check the latest paper from our group on resolving the impact of polar side chains on electronic and structural properties of donor-acceptor (DA) interfaces of organic semiconductors: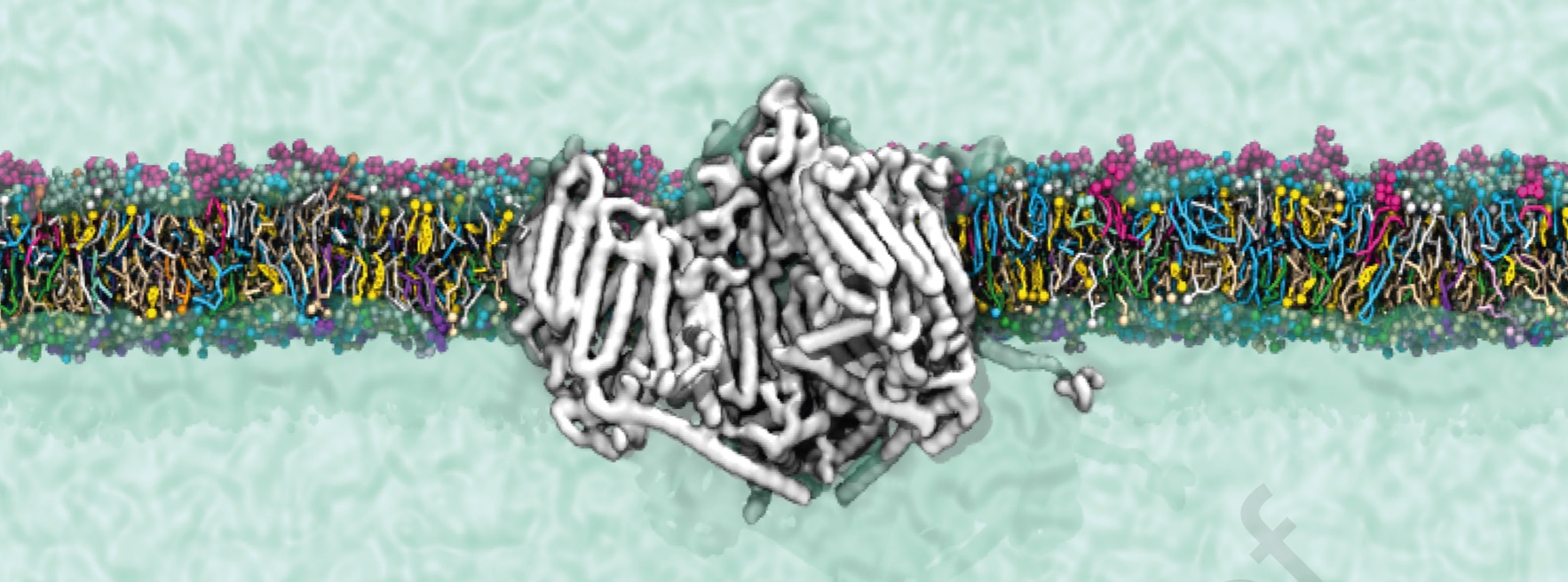 Interested in mechosensitive channels or protein-lipid interplay ?
Interested in mechosensitive channels or protein-lipid interplay ?