Modeling Lipid Particles
- Details
- Last Updated: Wednesday, 15 December 2021 08:46

Low-density and high-density lipoproteins (LDL/HDL) consist of a mixture of components including phospholipids, triglycerides, cholesterol, and associated proteins. The Martini model has proven useful to study the structure and dynamics of these particles [1-3].
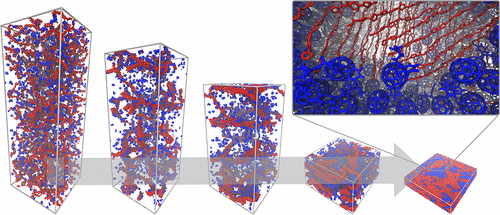
Other lipid based particles that have been simulated with Martini include a large variety of nanodiscs i.e., lipid bicelles stabilized by scaffolding proteins or DNA [8], or styrene-maleicacid copolymers [6] (figure), as well as liprotides [5] and complex nanoemulsions for vitamin delivery [9].
In addition, the process of triglyceride nanodroplet formation is being addressed with Martini [7], as well as the formation and transfection of lipid/DNA gene delivery vectors (lipoplexes) [10].
To efficiently build lipid-based nanoparticles, a number of dedicated tools are available [4,11].
- [1] A. Catte, J.C. Patterson, D. Bashtovyy, M.K. Jones, F. Gu, L. Li, A. Rampioni, D. Sengupta, T. Vuorela, P. Niemela, M. Karttunen, S.J. Marrink, I. Vattulainen, J.P. Segrest. Structure of spheroidal HDL particles revealed by combined atomistic and coarse grained simulations. Biophys. J., 94:2306-2319, 2008.
- [2] T.A. Vuorela, A. Catte, P.S. Niemela, A. Hall, M.T. Hyvonen, S.J. Marrink, M. Karttunen, I. Vattulainen. Role of lipids in spheroidal high density lipoproteins. PLoS Comp. Biol., 6:e1000964, 2010.
- [3] T. Murtola, T.A. Vuorela, M.T. Hyvonen, S.J. Marrink, M. Karttunen, I. Vattulainen. Low density lipoprotein: Structure, dynamics, and interactions of ApoB-100 with lipids. Soft Matter, 7:8135-8141, 2011.
- [4] Y. Qi, H.I. Ingólfsson, X. Cheng, J. Lee, S.J. Marrink, W. Im. CHARMM-GUI Martini Maker for coarse-grained simulations with the Martini force field. JCTC, 11:4486–4494, 2015. abstract
- [5] J.N. Pedersen, P.W.J.M. Frederix, J.S. Pedersen, S.J. Marrink, D. Otzen. Role of charge and hydrophobicity in liprotide formation: a molecular dynamics study with experimental constraints. ChemBioChem, 19:263–271, 2018. doi:
- [6] M. Xue, L. Cheng, I. Faustino, W. Guo, S.J. Marrink. Molecular Mechanism of Lipid Nanodisk Formation by Styrene-Maleic Acid Copolymers. Biophys. J., 115:494-502, 2018. doi:10.1016/j.bpj.2018.06.018.
- [7] X. Prasanna, V.T. Salo et al. Seipin traps triacylglycerols to facilitate their nanoscale clustering in the endoplasmic reticulum membrane. PLoS Biol 19(1): e3000998, 2021. https://doi.org/10.1371/journal.pbio.3000998
- [8] V. Maingi, P.W.K. Rothemund. Properties of DNA- and Protein-Scaffolded Lipid Nanodiscs. ACS Nano 15:751–764, 2021. https://doi.org/10.1021/acsnano.0c07128
- [9] N. Machado, B.M.H. Bruininks, ..., S.J. Marrink, P.C.T. Souza, P.P. Favero. Complex nanoemulsion for vitamin delivery: droplet organization and interaction with skin membranes. Nanoscale, 2021, online. https://doi.org/10.1039/D1NR04610A
- [10] B.M.H. Bruininks, P.C.T. Souza, H. Ingolfsson, S.J. Marrink. A molecular view on the escape of lipoplexed DNA from the endosome. eLife, 9:e52012, 2020. doi.org/10.7554/eLife.52012
- [11] L.R. Kjølbye, L. De Maria, T.A. Wassenaar, H. Abdizadeh, S.J. Marrink, J. Ferkinghoff-Borg, B. Schiøtt. General Protocol for Constructing Molecular Models of Nanodiscs. J. Chem. Inf. Model. 61 (6), 2869–2883, 2021. https://doi.org/10.1021/acs.jcim.1c00157